Abstract
The evolution of venoms is the story of how toxins arise and of the processes that generate and maintain their diversity. For animal venoms these processes include recruitment for expression in the venom gland, neofunctionalization, paralogous expansions, and functional divergence. The systematic study of these processes requires the reliable identification of the venom components involved in antagonistic interactions. High-throughput sequencing has the potential of uncovering the entire set of toxins in a given organism, yet the existence of non-venom toxin paralogs and the misleading effects of partial census of the molecular diversity of toxins make necessary to collect complementary evidence to distinguish true toxins from their non-venom paralogs. Here, we analyzed the whole genomes of two scorpions, one spider and one snake, aiming at the identification of the full repertoires of genes encoding toxin-like proteins. We classified the entire set of protein-coding genes into paralogous groups and monotypic genes, identified genes encoding toxin-like proteins based on known toxin families, and quantified their expression in both venom-glands and pooled tissues. Our results confirm that genes encoding toxin-like proteins are part of multigene families, and that these families arise by recruitment events from non-toxin genes followed by limited expansions of the toxin-like protein coding genes. We also show that failing to account for sequence similarity with non-toxin proteins has a considerable misleading effect that can be greatly reduced by comparative transcriptomics. Our study overall contributes to the understanding of the evolutionary dynamics of proteins involved in antagonistic interactions.
Issue Section: INTEGRATING PERSPECTIVES ON ANIMAL VENOM DIVERSITY\
Lead author Dr. Ricardo C. Rodríguez de la Vega tells us more about the research and goals behind this work:
What originally interested you in this area of research, be it venoms or intragenome diversity of toxin-like gene families?
Both. My main scientific interest is how reciprocal selection in antagonistic biotic interactions is recorded at the genome level. The idea here is that recurrent interactions between species generate co-evolutionary dynamics such that, as one species evolves, selective pressures on the other change and vice-versa. This reciprocal selection would naturally ended up changing the genotypes of the interacting species, However, this co-adaptation signal can be difficult, even impossible, to read as multiple other selection pressures and random processes leading to fixation (drift) also shape the genomes. In my view, antagonistic interactions generate evolutionary battlefields with deployed “arms” that may be anything from behavioural to molecular traits. In a simple case where “effector” molecules produced by one organism are delivered into another organism targeting “receptor” molecules reducing its fitness, and insofar as these molecules have simple genetic bases, this can offer a direct link between identifiable genomic features and reciprocally selected multi-organism phenotypic outcomes.
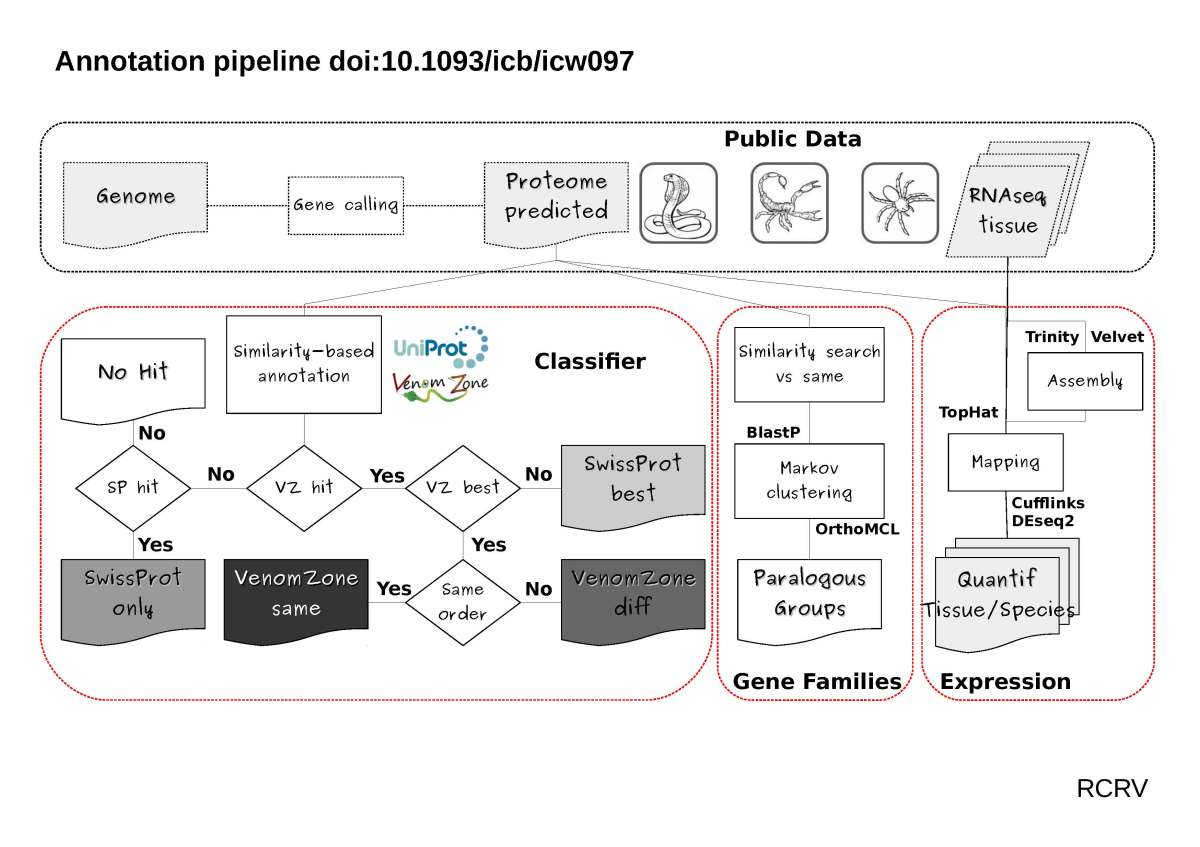
I am convinced that animal venoms provide many parallel examples of such simple and tractable case. Yet, the systematic study of these processes requires, of course, the reliable identification of molecules mediating the antagonistic interaction. Enters thus the problem of identifying the “effectors” of these interactions. In the case of animal venoms, the effectors, aka toxins, are the venom components whose selected function has been shaped by the action they have on ecologically relevant foes. Ribosomally synthesized toxins conform multigene families in venomous animals, understanding how these families arise is of utmost importance to understand the evolution of venom function. Until very recently toxins were thought to mainly originate from recruitment events of proteins with intra-organismal selected function, be it after gene duplication followed by neo- or sub-functionalization, or by re-purposing the original gene once it is expressed in the venom producing organ.
Did you feel that there was something missing from the field that motivated this work?
Yes. The advent of high throughput sequencing and improvements in proteomic protocols have relieved the burden of painstaking characterization of individual toxins directly from the venoms. Indeed, it is possible to identify candidate toxins directly from the genes expressed in the venom producing organs or even in whole genome surveys, by comparing the translated sequences with those in toxin databases such as UniProt’s venom protein annotation program. However, the existence of genes homologous to bona fide toxins but with no selected function in the venom, poses the much real problem of distinguishing between toxin and toxin-like genes in simple similarity searches. Taking advantage of recently available whole genome sequences of several venomous animals, I asked two very simple questions: 1) can we distinguish genes encoding toxins from toxin-like proteins by similarity searches (i.e. annotation transfer)? and 2) what is the relative contribution of toxin and toxin-like encoding genes to their corresponding multi-gene families?
What new insights do we gain as a result of this work?
First, that properly done, similarity searches can distinguish between toxin and toxin-like encoding genes. Second, that for many toxin containing mutligene families, non-toxin homologs outnumber the bona fide toxins, Third, that merging toxin and non-toxin homologs mislead the interpretation of recruitment events and selection regimes.
What are the critical future direction(s) do you anticipate this field moving in?
Having demonstrated that toxins indeed belong to multigene families, but that most of their homologs are unlikely to have a selected function in the context of venom use is in line with more recent evidence of highly dynamic venom composition, not due to recent duplication and pervasive diversifying selection (the standard model of venom evolution), but rather to pervasive and differential gene losses and to the rapid turnover of genes to acquire venom gland expression (assuming these genes have/have had indeed a selected function in the context of venom use).
What did you personally find most interesting, fun, or rewarding about this research?
I found particularly rewarding to be part of what I consider an authentic coming to age volume on venom evolution.



















 Ms. Keisha S Lewis is a former K-12 Biology teacher in Trinidad & Tobago, West Indies. She has a Bachelor’s degree in Biochemistry and a Postgrad Diploma in Education. While teaching, she founded and managed an annual science fair for over 10 years, as well as developed websites for her classes. She also previously served as an Assistant Examiner for the Caribbean Examinations Council (CXC). She is now venturing into the worlds of social media management and science communication. Follow her on Twitter
Ms. Keisha S Lewis is a former K-12 Biology teacher in Trinidad & Tobago, West Indies. She has a Bachelor’s degree in Biochemistry and a Postgrad Diploma in Education. While teaching, she founded and managed an annual science fair for over 10 years, as well as developed websites for her classes. She also previously served as an Assistant Examiner for the Caribbean Examinations Council (CXC). She is now venturing into the worlds of social media management and science communication. Follow her on Twitter